DNA repair and mutagenesis are essential for the maintenance and plasticity of genomes in all organisms. Our group studies these fundamental processes in bacteria, where mutagenesis underlies the evolution of antibiotic resistance. When bacterial cells are exposed to DNA damage or antibiotic treatment they launch stress responses that increase their survival. Intriguingly, these responses also upregulate mutation rates, accelerating genetic adaptation. Our recent findings show that stochasticity, or "noise", plays key roles in these responses, modulating mutation rates and diversifying cell phenotypes to evade drug treatment. Our goal is to understand the molecular processes that regulate DNA damage responses, repair, and mutagenesis and how they determine cell fates.
To this end, we take a quantitative multi-disciplinary approach. We are developing fluorescence microscopy and microfluidics methods to measure protein function at the level of single molecules in individual cells. By combining super-resolution microscopy and single-molecule tracking, we can directly watch the movement of individual proteins inside living cells.
We are particularly interested in the consequences of gene expression noise and stochastic processes on the fidelity of genome maintenance. To resolve cellular heterogeneity, we design custom microfluidic devices for the observation and manipulation of single cells. Using these tools, we discovered that the activation of a bacterial DNA damage response is highly stochastic, causing cell-to-cell variation in mutation rates. Random phenotypic variation can therefore lead to heritable genetic changes. We are currently exploring how bacterial cell heterogeneity influences the efficacy of antibiotic treatment.
We also collaborate with groups at Oxford and internationally on a range of projects related to genome maintenance in bacteria and eukaryotes.
Single-molecule imaging in live cells
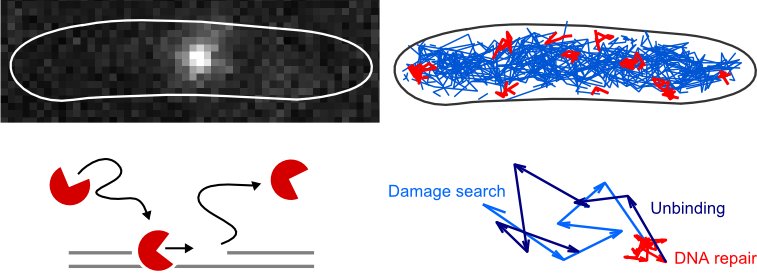
We are applying single-molecule fluorescence microscopy to directly observe individual proteins in living bacteria. One measurement of this type provides a wealth of quantitative information about molecular mechanisms at a single-cell level, such as the abundance and localizations of reaction sites, and the diffusion coefficients and dissociation constants of proteins and their complexes. This approach is very general and powerful, and continues to generate novel insights into the organization of DNA repair pathways, DNA replication, transcription, and chromosome organization.
Find more information in these publications: Stracy et al. Nature Communications 2016, Uphoff et al. DNA Repair 2014; Uphoff et al. PNAS 2013.
Microfluidics for single-cell analysis
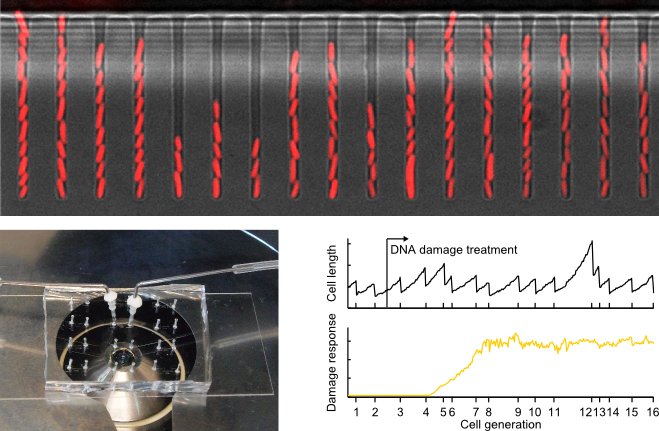 We use microfluidics to investigate how individual cells respond to DNA damage. The image shows E. coli cells growing inside microfluidic channels. Single cells at the bottom of each channel can be observed continuously over hundreds of generations. This allows us to follow response dynamics and measure phenotypic variation. These experiments inform about the regulation of DNA damage responses and cell fates.
Find out more information in these publications: Uphoff et al. Science 2016, Uphoff PNAS 2018
We use microfluidics to investigate how individual cells respond to DNA damage. The image shows E. coli cells growing inside microfluidic channels. Single cells at the bottom of each channel can be observed continuously over hundreds of generations. This allows us to follow response dynamics and measure phenotypic variation. These experiments inform about the regulation of DNA damage responses and cell fates.
Find out more information in these publications: Uphoff et al. Science 2016, Uphoff PNAS 2018
Real-time dynamics of mutagenesis
A central goal in genetics is to understand how mutation rates are regulated by the many genes that act in the creation or prevention of mutations. This fundamental questions remains despite decades of research, mostly because the methods to measure mutagenesis rely on accumulating mutations over time in large populations of cells.
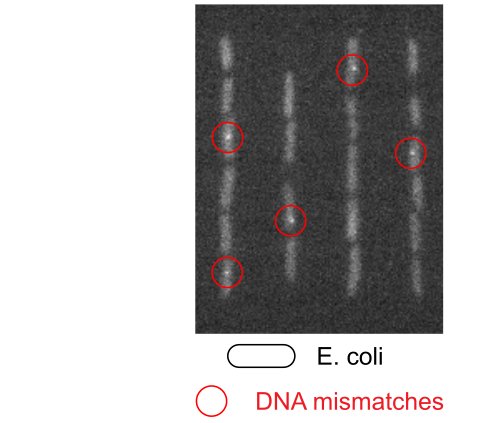
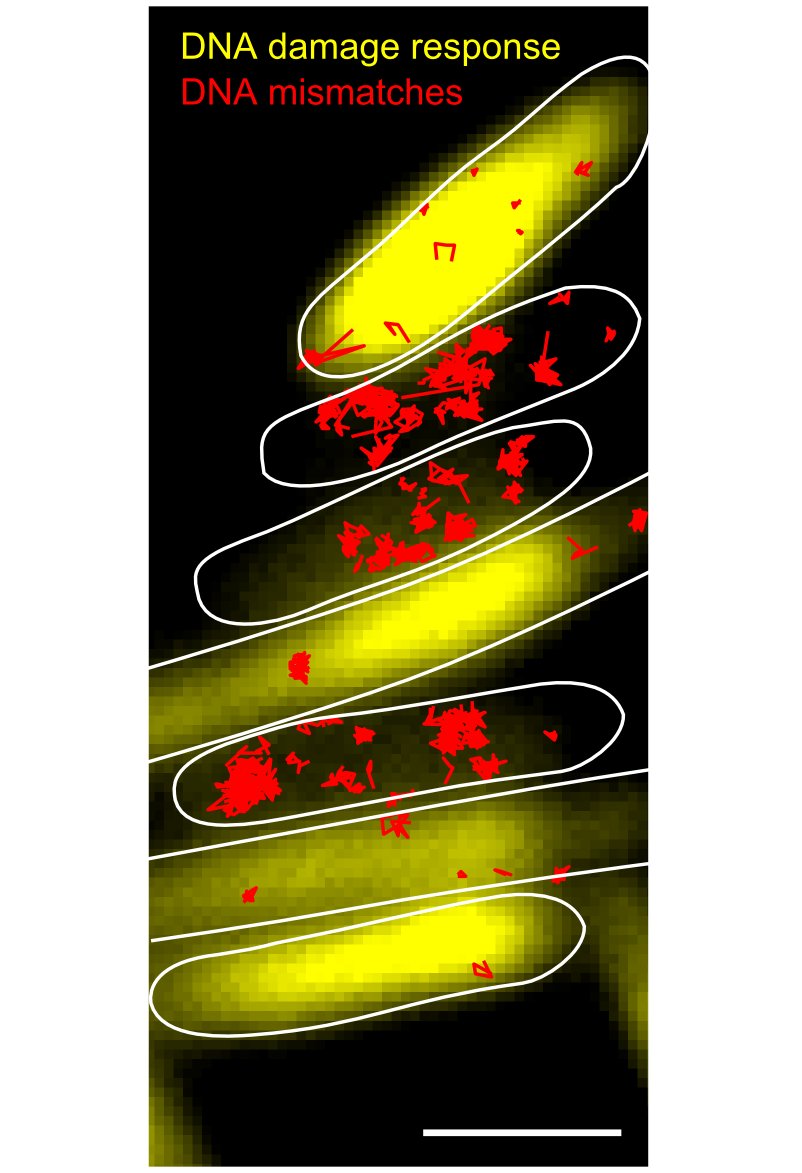
To overcome these limitations, we utilised a method to monitor mutagenesis in real-time in individual cells. With microscopy, it was possible to detect nascent DNA mismatches – the precursors of mutations – as fluorescent spots within growing and dividing cells. For the first time, the study revealed real-time changes in mutation rates after exposure to DNA damage. Surprisingly, mutagenesis occurred in a distinct pulse that was shaped by the expression of alternative genome maintenance pathways.
The discovery addresses the long-standing question what determines the choice between repair or tolerance of DNA damage, and how this choice affects mutagenesis. The results show that the alternative pathways follow a distinct chronology, where mutagenic damage tolerance is required for initial cell survival before the balance shifts in favour of accurate repair pathways. Furthermore, cell-to-cell heterogeneity in gene expression was found to modulate mutation rates. The general approach also revealed the dynamics of mutagenesis in response to antibiotic treatment.
Find out more information in this publication: Uphoff PNAS 2018.
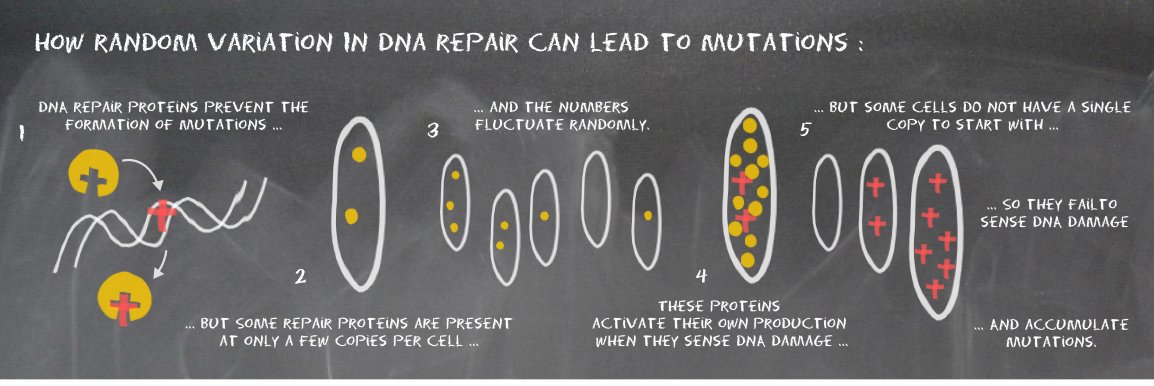
A link between mutagenesis and random variation in DNA repair
At the molecular level, all biological processes are subject to chance, governed by random encounters of molecules within cells. This means that even cells with identical genetic makeup will show random variation in their behaviour. We investigated whether such variation might affect the formation of mutations in the genome. The surprising results have now been published in Science.
We revisited the adaptive response to DNA methylation damage that was discovered in the bacterium Escherichia coli back in 1977, but using modern super-resolution fluorescence microscopy and single-cell microfluidic imaging. To our surprise, we found that the Ada protein required to sense DNA methylation damage was present at extremely low quantities in cells before damage. Cells had on average only a single copy of Ada, and a significant fraction of the cells had none at all. How many copies each cell contained was purely due to chance – or bad luck for those cells that had none. It turned out that the cells with no copies did not sense the presence of DNA damage. They were thus unable to activate the adaptive response that would allow them to repair the damage.
What are the consequences of this random variation for cells? Using a new method to visualize mutations under a microscope, we found that failure to activate the DNA damage response lead to the formation of mutations. The long-standing dogma in biology that genetic heterogeneity causes phenotypic heterogeneity is therefore also true in reverse: The capacity of cells to repair their DNA fluctuates randomly, and this phenotypic variation can lead to genetic changes.
Find out more information in this publication: Uphoff et al. Science 2016
Super-resolution microscopy method development
Super-resolution microscopy by single-molecule localization, such as STORM or PALM, employ photoswitching to image a small subset of molecules at a time while the majority of fluorophores reside in a non-fluorescent state. Thus isolated emitters can then be localized with high precision. The entire set of localizations acquired over the course of a movie forms a super-resolution image. Extension of this approach allows tracking the movement of individual molecules to directly monitor their activities in living cells.
We develop fluorescence microscopes that maximize detection sensitivity, mechanical stability, and flexibility for our different applications. These hardware developments are accompanied by a suite of custom data analysis software that we continuously improve and modify for each project. We also explore ways for fluorescent labelling of proteins inside live cells.
Find more information in these publications: Banaz et al. J Phys D 2018; Wegel et al Sci Reports 2016; Uphoff et al J Vis Exp 2014; Crawford et al. Biophys J 2013; Uphoff et al. PNAS 2013; Holden et al. Biophys J 2010; Holden et al. Nat Methods 2011.

Chromosome maintenance in eukaryotic cells
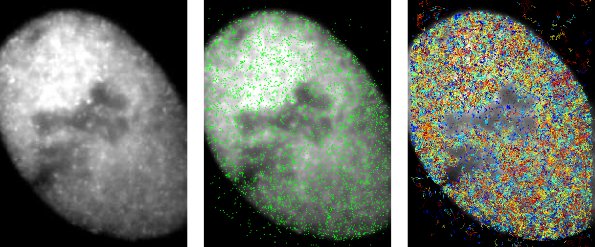
We are also interested in DNA repair and chromosome maintenance in eukaryotes. This image shows localisations of individual cohesin (Scc1) proteins that are bound to DNA (in green) and the tracks of molecules diffusing in the nucleoplasm (in rainbow colours).
Find out more about our study on the cohesin loader protein Scc2/Nipbl: Rhodes et al. bioRxiv 2017.